Iron Homeostatis and Cytoplasmic Control of Gene Expression
Iron. We all need it for many general cellular functions. When you don’t have enough Iron, your body cannot carry out those functions. On the flip side, you find that in high concentrations iron is toxic to the body. It is therefore critical that organism’s have a finely tuned system for regulating intracellular iron concentration.
Iron is used in many proteins as a cofactor, this includes oxygen transport proteins, hemoglobin and myoglobin. Iron plays an important role in oxygen transport and energy production, therefore it is not surprising that a shortage of iron in humans results in anemia, giving an over all feeling of weakness.
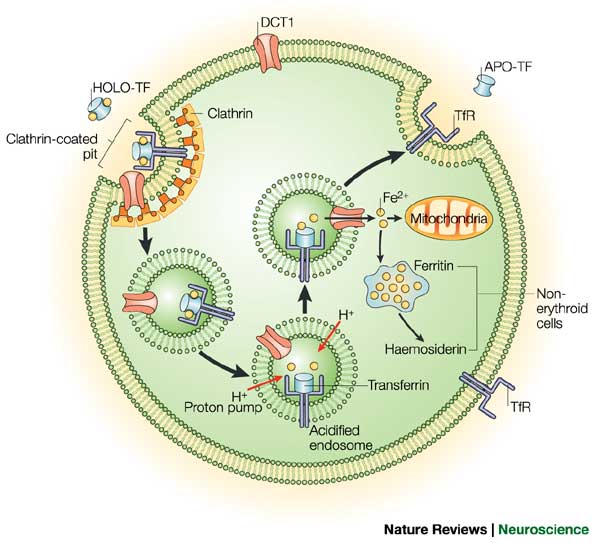
Two proteins, while not the only players in the regulation of iron homeostasis, provide an excellent and well studied example of regulation of gene expression post transcriptionally and in the cytoplasm. Let me introduce the players: Ferritin and the Transferrin receptor. Iron has extremely low solubility and is carried around by a third molecule (not discussed today) transferrin, and is taken up by the cell by the transferrin receptor, and stored by the cell in ferritin.
The levels of each of our two key players are regulated acording to the levels of iron in the following ways. When iron levels are high, there is an increase in Ferritin by increasing the Ferritin mRNAs affinity for the ribosome, therefore ensuring that protein gets made at increased levels, and excess iron gets stored. At the same time the levels of the Transferrin receptor are decreaeses so that there is less iron being taken up by the cell. This is regulated by regulating the stability of the Transferrin receptor mRNA.When iron levels are low then Ferritin levels are decreased so that less iron is stored, and the levels of Transferrin Receptor are increased by increasing the stability of its mRNA, and thus increasing the ability of the cell to take up more iron. These two proteins are differentially regulated by the same proteins. Let us explore how this is achieved.
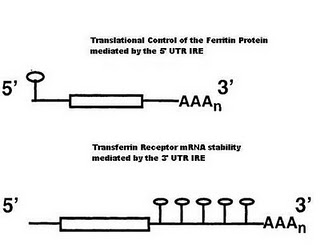
Iron Responsive Elements
In the 5′ UTR (untranslated region) of the Ferritin mRNA a stem loop structure that forms via base pairing interactions in the stem. The loop region contains a particular sequence of bases, which can be bound by IBP (Iron binding proteins).
In the 3′ UTR of the transferrin receptor are 5 of these stem loop IRE, which can bind the same IBP. These IRE’s are key to understanding the regulation of iron homeostasis.
Regulation of Ferritin
Ferritin translation is regulated by iron binding proteins. Such proteins are also RNA binding proteins that can recognize a specific hair-pin structured that is formed in the 3′ UTR of the Ferritin mRNA (its IRE). The affinity of the IBP for the IRE is controlled by the levels of iron in the cell. In iron deficient cells, the concentration of iron is too low to bind the IRP. In the absence of bound Iron these proteins are tightly bound to the Ferritin IRE and prevent the ribosome from initating translation.
Regulation of Transferrin Receptor
In the mRNA of the Transferrin receptor the 3′ UTR is 2.6 kb long and contains 5 IRE, whos sequence and structure is similar to the IRE found in the 5′ end of the ferritin mRNA. In the presence of low iron concentrations, the stability of the transferrin mRNA is increased by IBP, which bind to the IRE in the 3′ end. It is thought that such bound proteins prevent the receptor mRNA from entering the mRNA degredation pathway.
Experimental Evidence[![][5]][5]
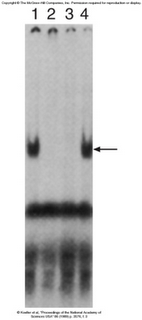
Gel Shift assays have demonstrated that the same protein that binds the TfR (Transferrin receptor) also binds the Ferritin IRE. A labled 3′ TrF mRNA was used in each of the lanes. Lane 1 has only cytoplasmic extract with no cold competitor added. We can see that there is an increase in the molecular weight of the labled mRNA, therefore showing that something has bound to the probe. Lane two contains a cold TrF mRNA, which is able to chase away the shift, therefore demonstrating that some protein is binding specifically to the TfR mRNA. Lane 3 shows that cold Ferritin can achieve the same thing, while lane 4 shows that in the presence of a cold non-specific competitor, specific binding still occurs.
The proteins that bind the IRE’s have been pulled down by UV cross linking, and demonstrated by Gel shift again. I wont be discussing this further because I am tired.